4749
Sparse DCE-MRI using a Temporal Constraint Learned from Clinical Data1Electrical Engineering, University of Southern California, Los Angeles, CA, United States, 2Radiology, University of Southern California, Los Angeles, CA, United States
Synopsis
Dynamic contrast enhanced MRI has benefitted substantially from developments in sparse sampling and constrained reconstruction. Thus far, temporal constraints have proven to be the most powerful. In this work, we explore the use of temporal dictionaries that are learned from a clinical database. We demonstrate that this method provides improved reconstruction quality compared to state-of-the-art TK-model-based constraints or low-rank constraints. The inclusion of spatial information while constructing dictionaries is also explored.
INTRODUCTION
Dynamic contrast enhanced (DCE) MRI
involves intravenous injection of a T1-shortening contrast agent
followed by the repeated acquisition of T1 weighted images providing
measurements of signal enhancement as a function of time1. The
dynamic anatomic images are then utilized to estimate tracer-kinetic (TK)
parameters such as vascular permeability and fractional plasma volume for each
voxel2. Standard methods are unable to simultaneously provide high
spatiotemporal resolution and volume coverage3. DCE has experienced
major improvements through the use of sparse data sampling4.
Parallel imaging (PI) and constrained reconstruction techniques have all been
used to provide substantial improvements in resolution and coverage5-7.
Recently, a model-based reconstruction that imposes TK model as a consistency
constraint was developed for DCE imaging and has enabled the highest under-sampling
rates to date8. However, this method may suffer from model
inconsistencies and inaccurate arterial input function extraction. Here, we investigate dictionary
learning (DL) algorithm that learns temporal constraint from clinical DCE-MRI
data. We found this technique provides improved performance compared to global
low rank (GLR)9 or model consistency (MOCCO)8 constraint.
METHODS
DL involves finding a basis set that is optimal for a specific set of signals to provide the sparsest possible representation for that particular signal10. The reconstruction problem can be expressed as the following optimization:
where are dynamic anatomic images, are the coil-sensitivities estimated using sum-of-squares coil combination, is the under-sampled space data with representing the -space coordinates, denoting the image domain spatial coordinates, and are the coil and time dimensions, is the under-sampling Fourier transform, is the learned dictionary, is the sparse representation, counts the number of non-zero elements, and is an operator that extracts 3D patches as a column vector. The optimization problem is non-convex and can be solved by splitting into two simpler subproblems. Iteratively solving these two subproblems will yield an approximate solution. Results were compared against GLR, MOCCO, and the fully-sampled reference. A fixed number of iterations was empirically chosen for both DL and GLR based on convergence behavior in retrospective under-sampling studies.
Dictionary Training. First, we assume is fixed while and are free variables. This step finds a dictionary that can sparsely represent the dataset using k-SVD algorithm10. The trained dictionary is later used to sparsely code the dataset using orthogonal matching pursuit (OMP)11.
Data Consistency. The second step is data consistency, in which is the only free variable. In this step, we insert the measured data at the sampling locations.
The datasets used in this study were from brain tumor patients receiving a routine brain MRI with contrast on a clinical 3 Tesla scanner at our institution. A 3D cartesian fast SPGR sequence with field of view (FOV): ; spatial resolution: ; temporal resolution: 5 seconds; 50-time frames; 8 receiver coils; flip angle; 1.3 ms echo time; and 6 ms TR was used. A dictionary of 800 elements was trained using 768000 patches from 20 fully-sampled datasets. This dictionary was then used to reconstruct 15 test datasets (retrospectively down-sampled).
RESULTS AND DISCUSSION
Reconstruction time was: GLR ~0.55 minutes for 50 iterations; MOCCO ~5.33 minutes; DL ~23.74 minutes for training and ~7.15 minutes for reconstruction with 20 iterations. The major computational burden is the sparse coding step, which takes ~ 6.67 minutes for 20 iterations.
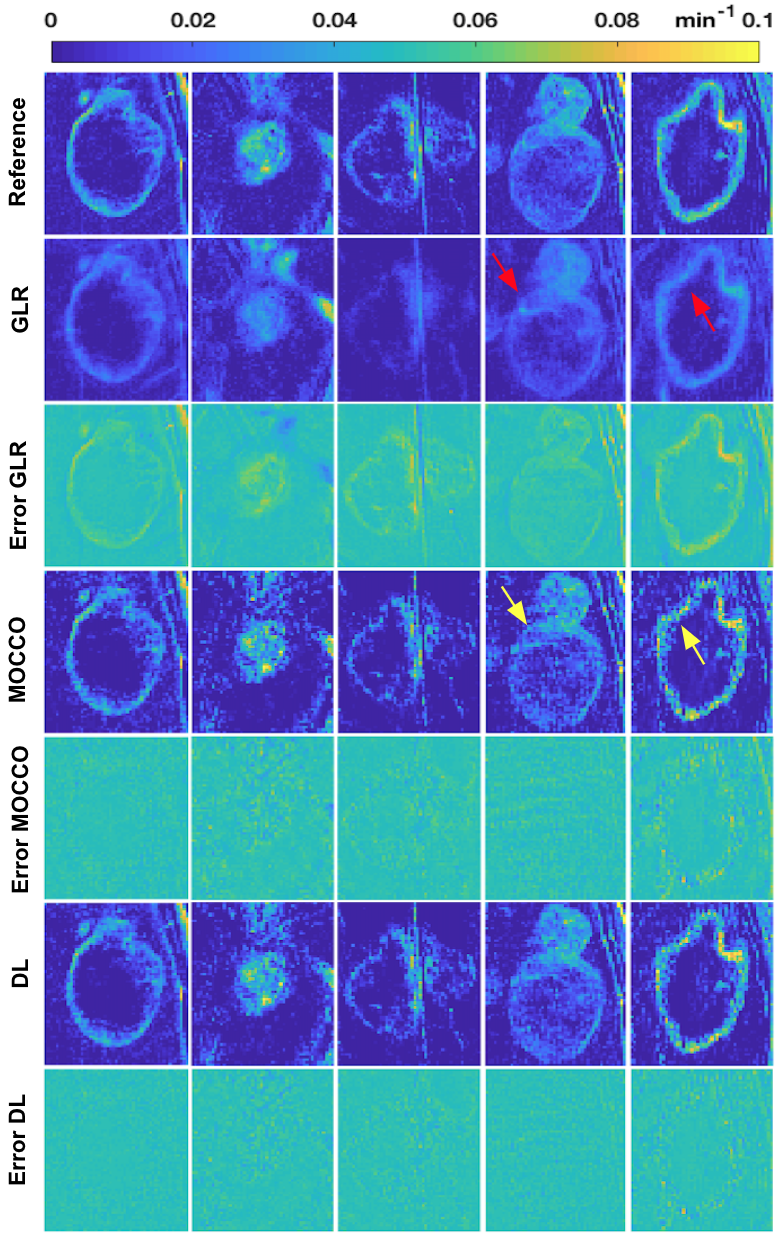
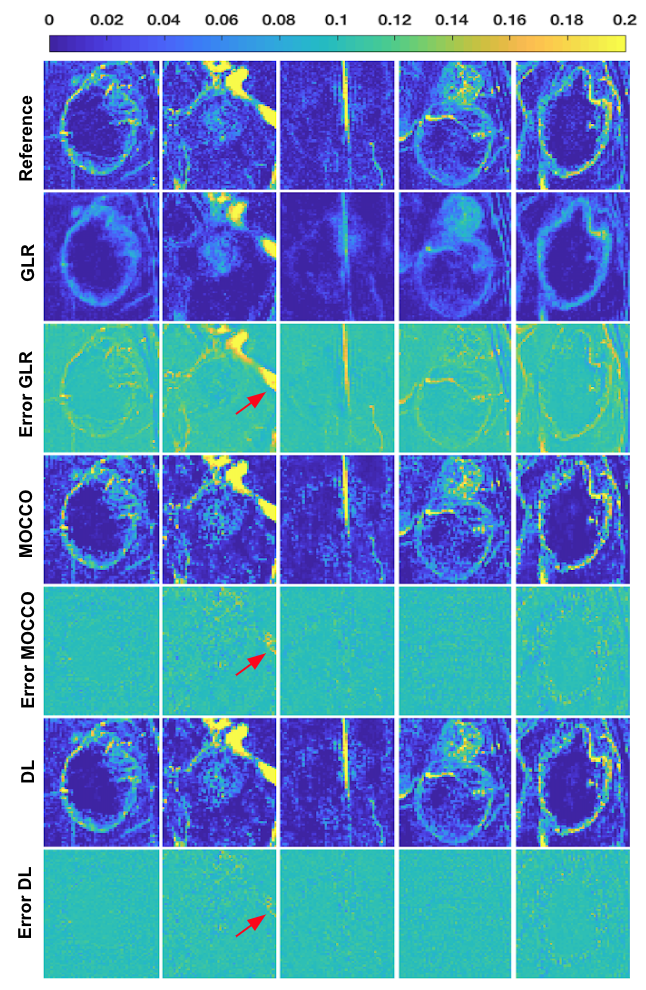
Figure 1 contains tumor ROI maps for 5 representative tumors (out of 15) for an under-sampling rate of 40 in the testing dataset. Figure 2 shows matching tumor ROI maps. TK maps reconstructed from GLR appear spatially smooth and those from MOCCO show a higher noise level.
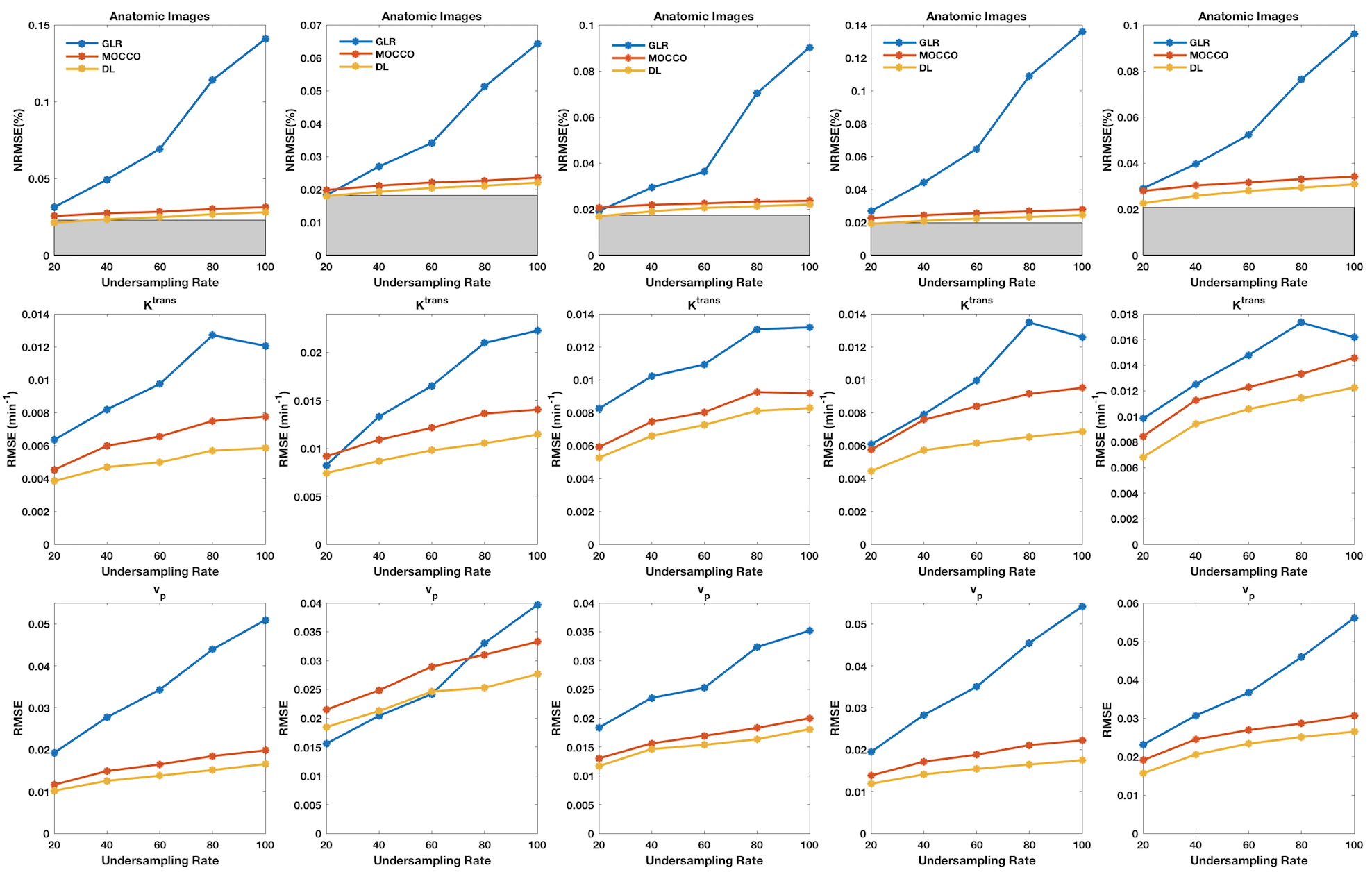
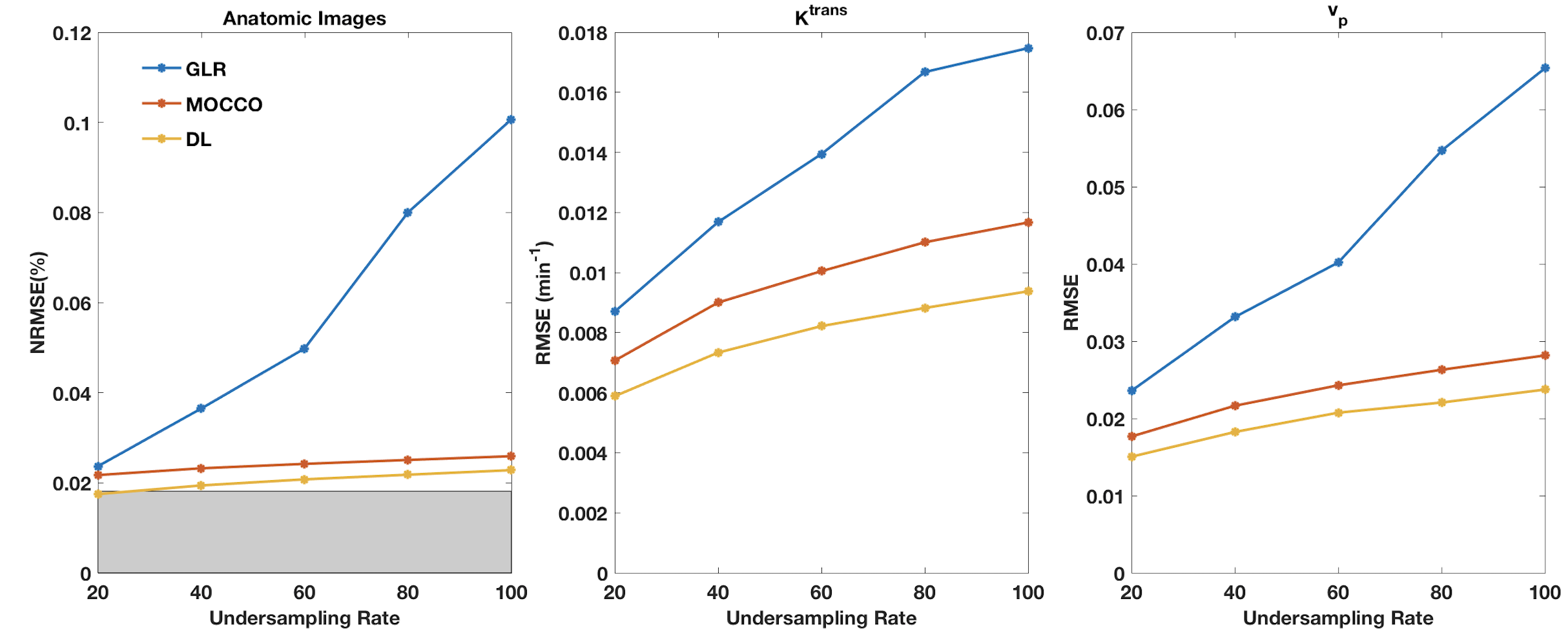
Figure 3 compares the performance as a function of under-sampling. Each column corresponds to the tumors shown in Figs. 1 and 2. DL consistently provided lower NRMSE compared to the alternatives that were studied. Figure 4 shows the average RMSE for all 15 tumors in the test dataset. The improvement of DL with respect to GLR is significant, whereas the improvement with respect to MOCCO is minimal.
We also explored the use of spatio-temporal dictionaries
(not shown here), and these provided significant denoising. This warrants
further exploration along with a comparison to previous spatio-temporal
constraints.
CONCLUSION
Sparse DCE-MRI reconstruction using a learned temporal constraint from clinical data may outperform current temporally constrained methods. The proposed technique was evaluated on 15 brain tumor cases and provided superior performance in every case.Acknowledgements
We acknowledge National Institute of Health grant support (#R33-CA225400).References
1. Heye AK, Culling RD, Hernandez MD, et al. Assessment of blood–brain barrier disruption using dynamic contrast-enhanced MRI. A systematic review. Neuroimage Clin. 2014;6:262-274.
2. Sourbron SP, Buckley DL. Classic models for dynamic contrast‐enhanced MRI. NMR Biomed. 2013;26(8):1004-1027.
3. Planey CR, Welch EB, Xu L, et al. Temporal sampling requirements for reference region modeling of DCE-MRI data in human breast cancer. J Magn Reson Imaging. 2009;30(1):121-134.
4. Paldino MJ, Barboriak DP. Fundamentals of quantitative dynamic contrast-enhanced MR imaging. Clin North Am. 2009;17(2):277-289.
5. Tudorica LA, Oh KY, Roy N, et al. A feasible high spatiotemporal resolution breast DCE-MRI protocol for clinical settings. Magn Reson Imaging. 2012;30(9):1257-1267.
6. Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182-1195.
7. Lebel RM, Jones J, Ferre JC, et al. Highly accelerated dynamic contrast enhanced imaging. Magn Reson Med. 2014;71(2):635-644.
8. Guo Y, Lingala SG, Bliesener Y, et al. Joint arterial input function and tracer kinetic parameter estimation from undersampled dynamic contrast‐enhanced MRI using a model consistency constraint. Magn Reson Med. 2018;79(5):2804-2815.
9. Zhang T, Pauly JM, Levesque IR. Accelerating parameter mapping with a locally low rank constraint. Magn Reson Med. 2015;73(2):655-661.
10. Aharon M, Elad M, Bruckstein A. K-SVD: An algorithm for designing overcomplete dictionaries for sparse representation. IEEE Trans. Signal Process. 2006;54(11):4311-4322.
11. Tropp JA, Anna, and Gilbert C. Signal recovery from random measurements via orthogonal matching pursuit. IEEE Trans. Inf. Theory. 2007;53(12):4655-4666.
Figures