4544
Sparse Pre-Contrast T1 Mapping for DCE-MRI Calibration1Electrical Engineering, University of Southern California, Los Angeles, CA, United States, 2GE Healthcare, Calgary, AB, Canada
Synopsis
Quantitative DCE-MRI requires fast pre-contrast T1 mapping (scan time <3 min) with matching resolution and coverage. Recent advances in imaging have substantially improved resolution and coverage of DCE-MRI but without matched improvements in the pre-contrast T1 data. Here, we demonstrate a sparse T1 mapping method and characterize a tradeoff between data acquisition and T1 statistics, using a variable flip angle (VFA) approach and sparse Cartesian spiral sampling pattern, with image domain wavelet sparsity constraint. This method provides the necessary high-resolution whole-brain T1/M0 maps for DCE-MRI tracer kinetic analysis.
Introduction
Dynamic contrast enhancement (DCE)-MRI is a powerful imaging tool that can reveal the spatial distribution of vascular parameters, including permeability and plasma volume. However, widespread clinical application is limited by low spatial resolution, insufficient spatial coverage and long data acquisition. Recent studies have overcome these limitations by using compressed sensing and constrained reconstruction techniques1,2. There is an unmet need for resolution and coverage matched pre-contrast T1 mapping. Lebel et al. demonstrated that T1 mapping is possible using sparsely sampled variable flip angle (VFA) acquisition integrated with DCE-MRI2. Maier et al. demonstrated sparse T1 mapping estimation using total generalized variation (TGV) constraints3. However, relationship between data acquisition acceleration and reconstruction quality remains unresolved. In this work, we demonstrate a pre-contrast T1 mapping2, and we also characterize the trade-off between data acquisition time and T1 mapping accuracy.Method
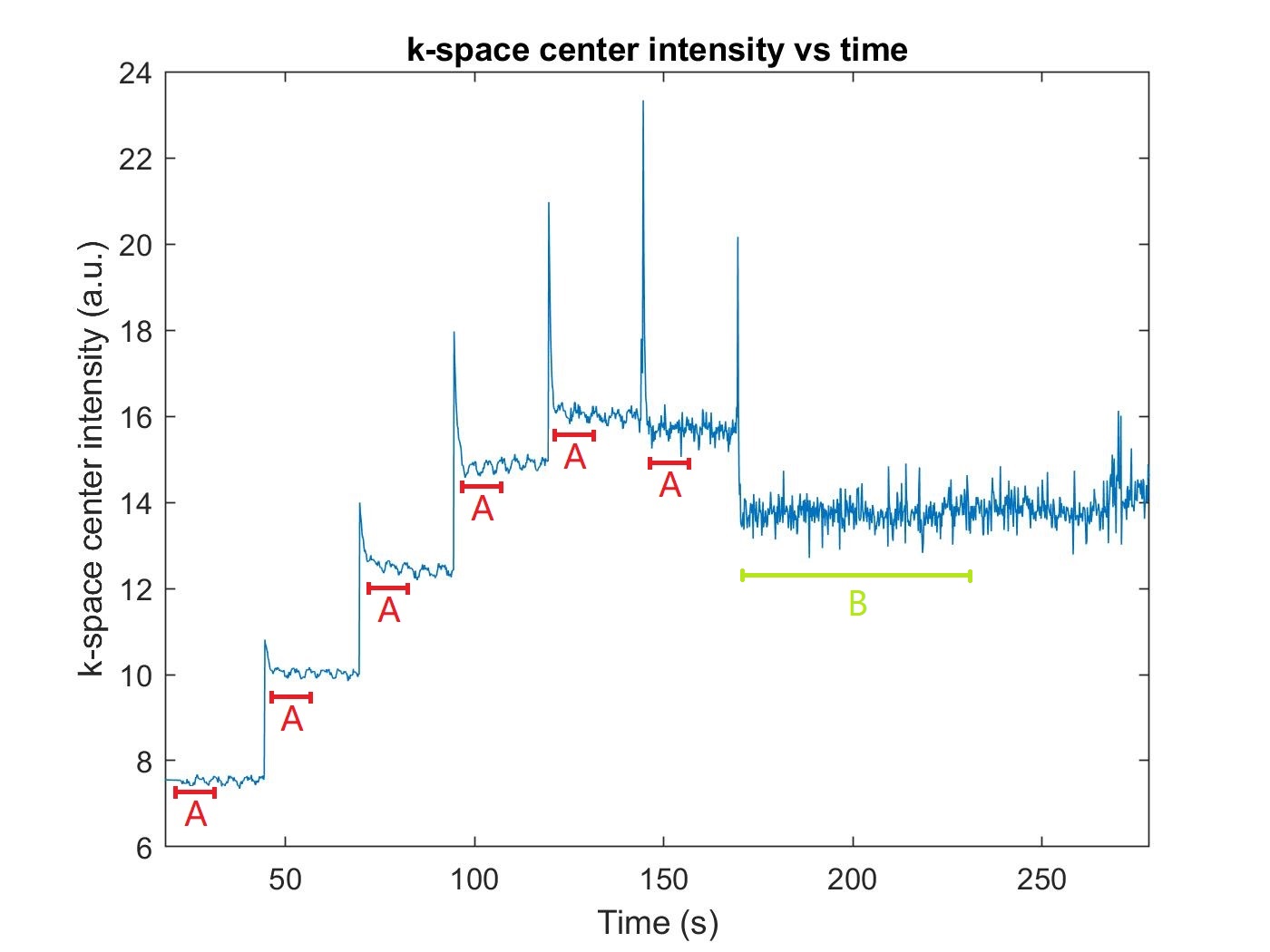
Data were acquired on a glioblastoma subject using a GE MR750 scanner with a 12-channel head coil. The vendor provided 3D spoiled gradient echo sequence was modified to include sparse undersampling. Six increasing flip angles each with 5000 TRs and a final flip angle with 20000 TRs were performed. Note that the last flip angle has a longer duration since it is used in the DCE scan and precedes the contrast injection. The flip angle increased from 1.5° to 15° logarithmically2. Other settings: 5 ms TR, 1.9 ms TE, 240×240×240 mm3 FOV, 2 mm slice thickness, and 256×240×120 matrix size.
Pre-contrast T1/M0 mapping is performed by solving the following constrained inverse problem2:
where the estimated anatomic image magnitude is
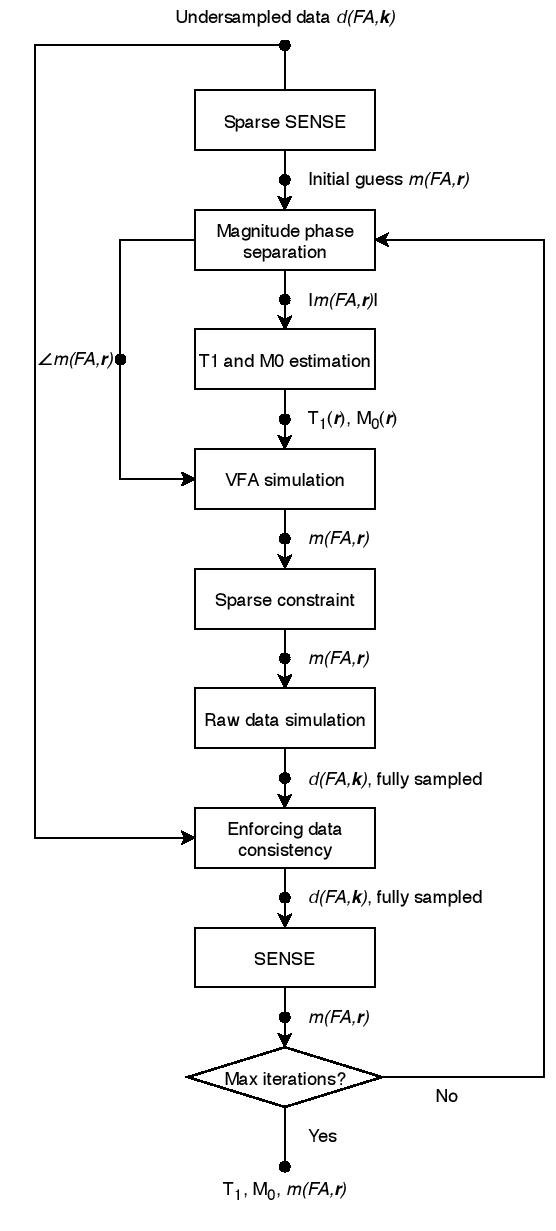
concatenates T1 and M0 into a vector, is the undersampled Fourier transform, is the coil sensitivity, is VFA images, converts T1 and M0 values into VFA images, is a sparsifying transform (e.g., wavelet), is measured -space data, and is a regularization parameter. This problem is solved using POCS iterations, which alternate between thresholding wavelet coefficients and forcing data consistency. A flowchart of this workflow is shown in Figure 1.
We used wavelet transform as the spatial sparsifier as it preserves subtle features as well as denoises data. was empirically chosen as 0.3 and remained constant in all experiments. To avoid model failure impacting the estimation process, we impose both non-negative and non-infinite constraints on each pixel’s T1 and M0 values.
We explored T1 mapping accuracy as a function of the number of TR periods included at each flip angle. Faster acquisitions were simulated by discarding samples at each flip angle, as illustrated in Figure 2. We measure and report the error in T1 and M0 maps as functions of the amount of retained data.
Results
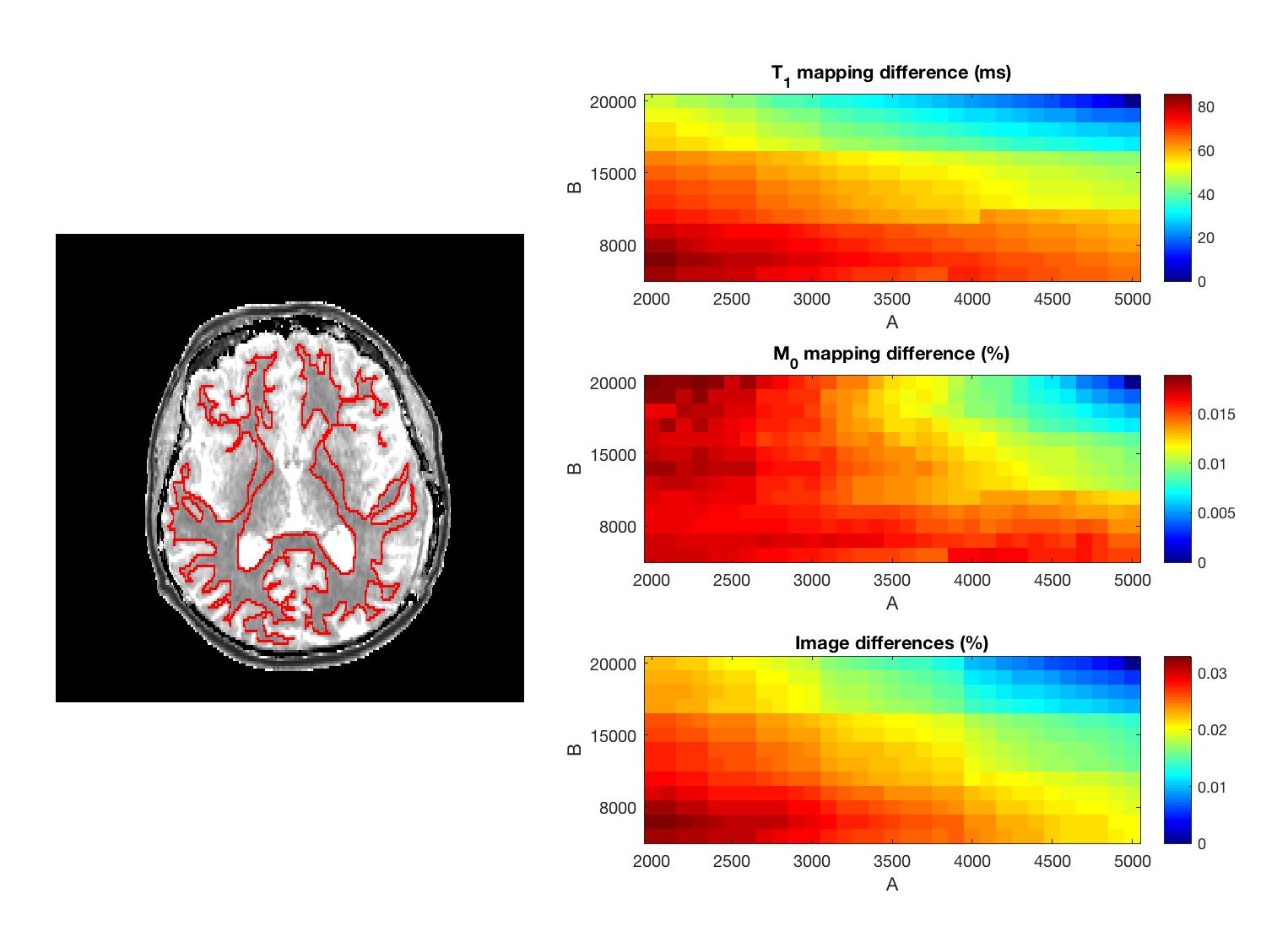
Figure 3 shows the ROI outline and difference maps for T1, M0, and anatomic images within the ROI. These maps were obtained by computing differences between results of (5000, 20000) setting and results of any (A, B) setting. Note that M0 difference decreases as B decreases when A is small. In addition, there was at most 85 ms T1 difference per pixel, and the differences in quantity of magnetization and image energy were 1.873% and 3.290% at most.
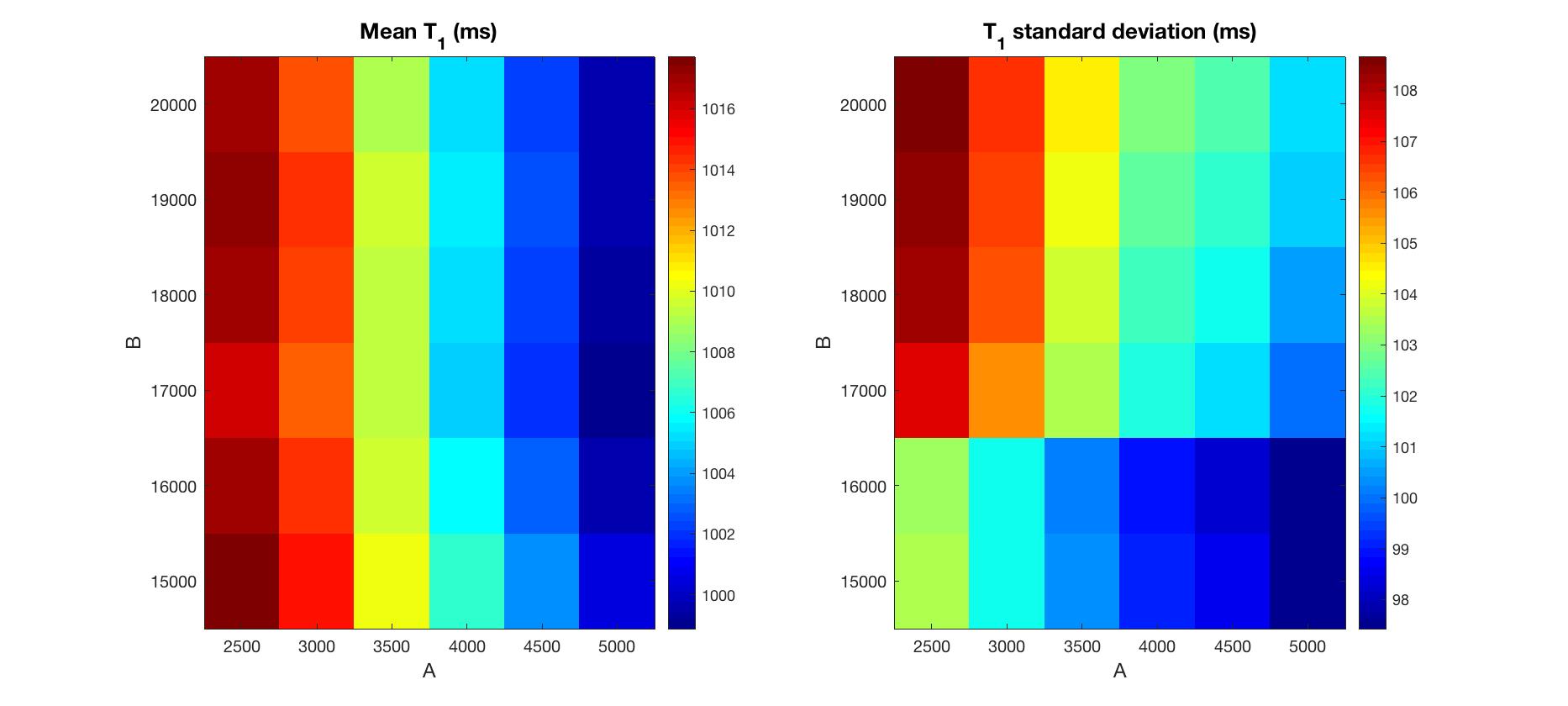
Figure 4 shows means and standard deviations of estimated T1 in normal white matter. T1 values were selected from the ROI in Figure 3. All results show similar mean T1. Interestingly, increasing A or decreasing B both result in a subtle decrease in standard deviation of T1.
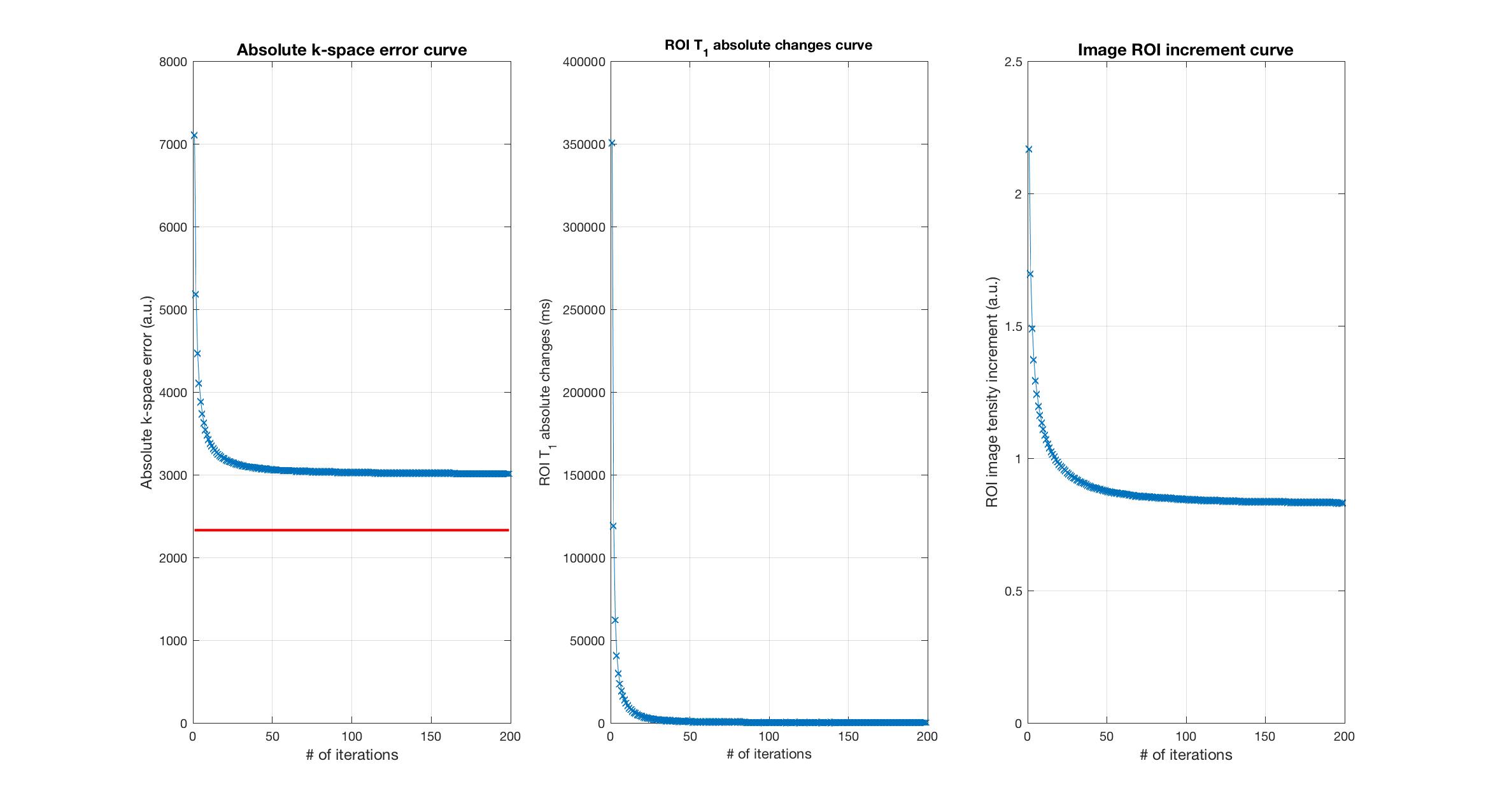
Figure 5 illustrates a -space error curve, a ROI T1 absolute change curve and a ROI image increment curve. Both T1 change and image increment became trivial after 80 iterations, while residuals in -space were quite significant.
Discussion
This approach enables adequate pre-contrast T1 estimation matched to the resolution and coverage of modern high-resolution whole-brain DCE-MRI in a reasonable scan time (<3 minutes), in stark contrast to a fully sampled VFA acquisition that would require more than 20 minutes. Despite different subsample settings, both T1 values mean and standard deviation were stable, and no substantial bias was observed. This indicates that the proposed approach is flexible and allows a trade-off between data acquisition time and T1 statistics.
Another important observation is significant -space residuals. One possible cause is that wavelet sparsity regularization can over-smooth images. One way to compensate for these residuals is to impose TGV constraints on vascular parameters directly3, or to enforce data consistency after each iteration (used in this work).
Conclusion
Sparse pre-contrast T1 mapping matched to high-resolution whole-brain DCE-MRI acquisition is feasible. There is a trade-off between data acquisition time and T1 accuracy.Acknowledgements
We acknowledge National Institute of Health grant support (#R33-CA225400).References
1. Guo, Y., Lebel, R. M., Zhu, Y., Lingala, S. G., Shiroishi, M. S., Law, M. and Nayak, K. (2016), High‐resolution whole‐brain DCE‐MRI using constrained reconstruction: Prospective clinical evaluation in brain tumor patients. Med. Phys., 43: 2013-2023. doi:10.1118/1.4944736
2. Lebel, R. M., Guo, Y., Lingala, S. G., Frayne, R., Nayak, K. S. "Highly Accelerated DCE imaging with integrated T1 mapping." Proc. ISMRM 25th Scientific Sessions, Honolulu, April 2017, p138.
3. Maier O., Schoormans J., Schloegl M., et al. Rapid T1 quantification from high resolution 3D data with model‐based reconstruction. Magn Reson Med. 2018;00:1–18. https://doi.org/10.1002/mrm.27502
Figures